Welcome to the Computational Modeling of Nanosystems course by Mario Barbatti.
Course presentation
Welcome to the enchanting world of Computational Modeling of Nanosystems! Get ready for a mind-blowing journey into the tiniest wonders of life! I’m your guide on this thrilling adventure, where you’ll become a molecular architect, crafting 3D models of the building blocks that make up everything around us.
In this Master’s level course, we’ll unlock the door to groundbreaking discoveries and innovation. From designing new drugs to predicting photochemical behavior, the possibilities are endless.
Sure, it won’t be a walk in the park, but we’ll take it step by step, and you’ll have a blast. Masterclasses, tutorials, and practical works were designed to drive you through the mathematics, physics, and chemistry we need in this endeavor.
So, aspiring molecular wizards, get ready to turn complex concepts into a playground of learning and growth. The adventure begins now. Are you ready? Let’s do this! 🚀
– Mario Barbatti
Course program
Our Computational Modeling of Nanosystems course is composed of three parts:
- Quantum mechanics
- Classical mechanics
- Statistical mechanics
Each part has four masterclasses followed by a few tutorials and practical work sections.
I will be in charge of the masterclasses. The practical work will count on the help of my colleague Vijay Chilkuri. Members of the Light & Molecules team will teach the tutorials.
Evaluation
The evaluation will be based on three tasks assigned to practical works (TP):
- A short literature review that you should write (TP2) (25%)
- A molecular dynamics program that you should code (TP3, TP4, and TP5) (50%)
- A seminar you should deliver (TP6) (25%)
The literature review and the seminar can be on the same topic. See TP2 below for instructions about how to choose a topic.
Although these tasks are scheduled to specific TPs, you can start working on them from day one after assigning your research topic.
You can discuss and work with colleagues, but the MD program and literature review are individual works you must do yourself.
For researching, writing, illustrating, coding, and any other tasks, you can (and must!) use generative AI (ChatGPT, BingAI, Bard, Midjourney, …) to help. However, you are the author. Any mistake, omission, or false information is not a machine’s fault; it’s yours!
Also, plagiarism is a deadly sin.
Course Schedule
| Date | Morning | Afternoon |
|---|---|---|
| Part I | ||
| 18-Sep | Lecture QM1 | Practical work TP1 |
| 25-Sep | Lecture QM2 | Tutorial TD1 |
| 02-Oct | Lecture QM3 | Tutorial TD2 |
| 09-Oct | Lecture QM4 | Practical work TP2 |
| Part II | ||
| 16-Oct | Lecture CM1 | Tutorial TD3 |
| 23-Oct | Lecture CM2 | Practical work TP3 |
| 06-Nov | Lecture CM3 | Tutorial TD4 |
| 13-Nov | Lecture CM4 | Practical work TP4 |
| Part III | ||
| 20-Nov | Lecture SM1 | Tutorial TD5 |
| 27-Nov | Lecture SM2 | Practical work TP5 |
| 04-Dec | Lecture SM3 | Tutorial TD6 |
| 11-Dec | Lecture SM4 | Tutorial TD7 |
| 08-Jan | Practical work TP6 |
Program of the Masterclasses
| I – QUANTUM MECHANICS | |
| QM1 – Intro to QM & Born-Oppenheimer approximation | |
| Wave functions and the Schrodinger Equation | |
| BO approximation: potential energy surfaces | |
| A math reminder | |
| The BO approximation in detail | |
| QM2 – Quantum chemistry | |
| BO approximation: TD perspective | |
| A note about molecular time | |
| Introduction to quantum chemistry | |
| QM3- Beyond BO approximation | |
| A bit of history: Born & Oppenheimer | |
| Nonadiabatic couplings and conical intersections | |
| Nonadiabatic dynamics | |
| QM4 – QM in context | |
| From quantum to classical nuclear motions | |
| Wave function decoherence and collapse | |
| Framing molecules in the Core theory | |
| II – CLASSICAL MECHANICS | |
| CM1 – Newton’s laws | |
| Mechanics of a particle | |
| Mechanics of a system of particles | |
| Reference frames (toward special relativity) | |
| EOM integration | |
| CM2 – Molecular mechanics: normal modes | |
| Harmonic approximation | |
| Normal modes | |
| Continuous medium | |
| Spin-Boson PES | |
| QM of normal modes | |
| Using the normal modes | |
| CM3 – Molecular mechanics: dynamics | |
| BO MD | |
| Fitted PES | |
| Model Hamiltonian | |
| Force fields | |
| QM/MM | |
| Continuum model | |
| Adiabatic x diabatic PES | |
| Photophysics in active environments | |
| CM4 – Hamilton and Lagrange formulations & MQCD | |
| Mixed-quantum classical methods | |
| Newton-X | |
| The cost of dynamics | |
| Lagrange’s equations | |
| Car-Parrinello MD | |
| Hamilton’s equations | |
| III – STATISTICAL MECHANICS | |
| SM1 – Principles of SM | |
| The Boltzmann picture | |
| Maxwell-Boltzmann statistics | |
| Particle distributions | |
| The Gibbs picture | |
| Thermostats | |
| SM2 – Monte Carlo algorithms, sampling techniques, and rates | |
| Monte Carlo & Metropolis | |
| Rate theory: Fermi, Marcus, emission | |
| SM3 – Fourier transform and spectrum simulations | |
| Statistical errors in MD | |
| Spectrum simulations | |
| Pyrene as case study | |
| SM4 – Machine learning | |
| Supervised ML: Dynamics simulations | |
| Unsupervised ML: Dynamics analysis |
Content of the Tutorials
| Tutorials |
| TD1 – Best practices for research (Rafael Mattos) |
| TD2 – Mathematics review (Mario Barbatti) |
| TD3 – Python basics (Rafael Mattos) |
| TD4 – Molecular dynamics and QM/MM Tutorial (Josene Toldo) |
| TD5 – DFT & TDDFT Tutorial (Saikat Mukherjee) |
| TD6 – Nuclear ensemble and surface hopping Tutorial (Saikat Mukherjee) |
| TD7 – Machine learning Tutorial (Matheus Bispo) |
Info about the Practical Works
| Practical works | Delivery date |
| TP1 – Software installation | |
| TP2 – Topic review writing | Before 20/12 |
| TP3 – PES coding | Before 30/10 |
| TP4 – MD coding | Before 20/11 |
| TP5 – Thermostat coding | Before 08/12 |
| TP6 – Seminars | 08/01 |
TP1 – Software installation
We will need several programs during the Molecular Modeling course. In TP1, we will install them on our laptops.
| Program | Function | Website |
|---|---|---|
| Anaconda | Python management + Jupyter | https://docs.anaconda.com/free/anaconda/install/ |
| VScode | Programming editor | https://code.visualstudio.com/download |
| WSL | Linux for Windows (Only for Windows laptops) | https://learn.microsoft.com/en-us/windows/wsl/install |
| MobaXTerm | SSH (Only for Windows laptops) | https://mobaxterm.mobatek.net/download.html |
| JMOL | Molecular visualization | https://jmol.sourceforge.net/download/ |
| MS Office | Text, worksheets, presenttations (Maybe it’s already installed?) | https://login.microsoftonline.com/login.srf?wa=wsignin1.0&whr=univ-amu.fr |
| Zotero | Literature management (Only for Windows) | https://www.zotero.org/download/ |
| LabPlot | Graphics | https://labplot.kde.org/download/ |
| Grammarly | Writing assistant | https://www.grammarly.com/desktop |
| Chrome | Browser (Likely it’s already there) | https://www.google.com/chrome/ |
| Lean library | Journal access (Only after installing Chrome!) | https://download.leanlibrary.com/aix-marseille-universite |
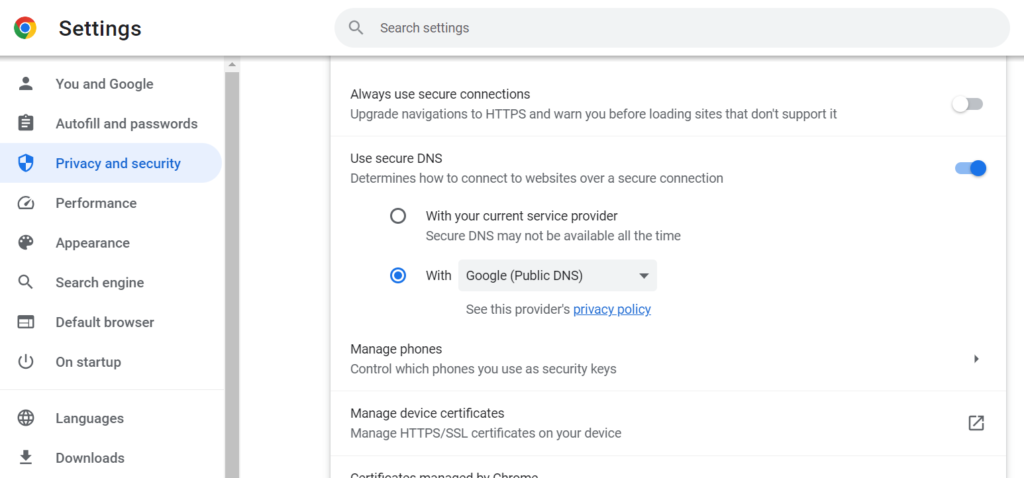
After installing Chrome, go to Settings > Privacy and Security> Security Change DNS to “Google (Public DNS).”
TP2 – Topic review writing
You will write a literature review and give a seminar (TP6) on a specific topic. The topic assignment is provided below.
| Topic | Student |
|---|---|
| Graphene oxide | M. Belay |
| Supercapacitors: perspective to energy storage | A. Diima |
| Cellulose nanomaterials | W. Perera |
| Nanoporous materials: applications in adsorption | J. Attupuram Joychan |
| Green nanotechnology | K. Duerme |
| Nanomaterials for electrocatalytic water splitting for hydrogen generation | L. Shaju |
| Quantum dots | R. Ahmad |
| Green synthesis of quantum dots | B. De La Toba Acevedo |
| Biologically inspired nano-assembly: 2D DNA origami | M. Ahmed |
| Nanotechnology for water and water waste treatment | M. Mohamed |
| Organic photovoltaics | E. Ali |
| Inorganic photovoltaics | M. Tahir Hafeez |
| Electrocatalysis in Zn-air Battery | N. Moustafa |
| Atmospheric photochemistry | C. Crîmpiță |
| Quantum computing | S. Mburu |
| Nanotechnology for carbon capture | Z. Farzizada |
| Graphene-Reinforced Aluminum Alloy Nanocomposites | A. Marterior |
| New developments in high-temperature supercondutivity | I. Islam |
| Chromophore development for sunscreen | L. Younes |
| Machine learning for predicting molecular properties | M. Karasek |
The review should focus on molecular aspects and, when pertinent, on molecular modelization features.
In terms of format, the review should:
- Be in a standard paper format (Abstract, Introduction, Middle sections, Conclusions)
- Be fully referenced and illustrated
- Have about 2000 to 3000 words
- Emphasize recent research (after 2020)
- Survey at least 15 references
You can discuss and share notes with a colleague writing on a similar topic, but the review is individual.
Your paper should:
- Introduce the topic and tell why it is relevant
- Briefly explain it
- Review the latest findings and developments
- Draw (and even speculate about) perspectives for the future.
We will evaluate:
- Quality of research (pertinence of the works surveyed)
- Quality of insights (your perspective on the topic)
- Quality of writing (we expect publication-level English. Grammarly is your friend, Use it!)
- Quality of formatting (the document should be beautiful, structured, and clean)
I suggest you use this template:
If you are uncomfortable writing in English, I strongly recommend Pinker’s The Sense of Style.
TP3, TP4, and TP5 – Molecular dynamics coding
We will code an analytical potential energy surface in Python and run microcanonical and canonical dynamics on it. This will be done in a Jupyter or Google Colab Notebook, which will be used in TP3, TP4, and TP5.
Your notebook should:
- Fully documented, clear, and understandable by someone else
- Run smoothly
You can work with other colleagues, but each must have their own notebooks, which must not be identical.
We will evaluate:
- Quality of coding
- Quality of the notebook documentation
- Quality of the insight delivered analyzing the data
If your programming skills are not up-to-date, take a look at the following materials:
For the evaluation, you should share your Jupyter notebook with the teacher responsible for the TP (it can be Dr. Vijay Chilkuri or myself). You can directly send us your .jpynb file. If you use Google Colab, it is enough to send us the link, but do not forget to make it available for anyone with the ink. (If you want to know more about sharing with Google Colab, check this video.)
The tasks and theory needed for these TPs are described in this document.
TP6 – Seminars
Now it’s your turn to teach. You should give a 15-minute seminar for your colleagues about the topic of your short review (TP2).
Your presentation should be about 10 min, with 5 minutes for questions.
We will evaluate:
- The clarity of your message
- Your answers to questions
- Quality of your slides
- Time management
Literature and textbooks
The course does not follow a single source, but it references many books, papers, and internet materials like essays and videos. The references are given during the lectures. Most of them can be downloaded from the link below (the password will be given in the first lecture).
https://amubox.univ-amu.fr/s/xXAiMZrDPb9RMRX
CNE Class of 2023