New Software Unveils Hidden Patterns in Nonadiabatic Dynamics.
In brief:
- Streamlined Analysis: ULaMDyn automates the preprocessing, dimensionality reduction, and clustering of NAMD datasets, simplifying the study of complex excited-state processes.
- Actionable Insights: This tool identifies critical molecular geometries and transitions, offering a deeper understanding of photochemical dynamics, such as conical intersections.
- Scalable and Flexible: ULaMDyn, designed for accessibility, supports standard and high-performance computing workflows and is adaptable to diverse molecular systems.
Our recent publication in Digital Discovery [1] introduces ULaMDyn, a Python-based open-source tool for analyzing nonadiabatic molecular dynamics (NAMD) simulations. By applying advanced unsupervised learning methods, ULaMDyn simplifies the interpretation of high-dimensional molecular datasets, enabling researchers to uncover critical patterns in complex excited-state dynamics.
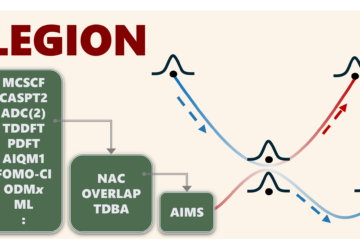
Nonadiabatic processes, which are crucial in photophysics and photochemistry, are notoriously challenging to analyze due to their intricate trajectories and high-dimensional data. ULaMDyn addresses this challenge by seamlessly integrating with the Newton-X platform and automating data preprocessing, dimensionality reduction, and clustering. This comprehensive workflow empowers researchers to identify key molecular geometries and transitions, streamlining the study of phenomena like conical intersections and ultrafast nonradiative decay.
ULaMDyn was created by Max Pinheiro Jr., with the contributions of many members of the Light & Molecules group, to modernize data analysis on the Newton-X platform.
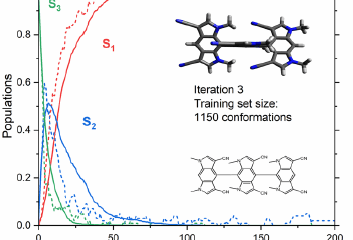
In our benchmark study, we applied ULaMDyn to analyze the dynamics of fulvene, a prototypical photoactive molecule. The tool efficiently identified critical structural features, such as bond stretching and puckering motions, that govern the molecule’s transition through conical intersections. Dimensionality reduction techniques like ISOMAP and clustering algorithms like K-Means allowed us to extract meaningful insights from large datasets, highlighting the relationship between structural variations and electronic state transitions.
Beyond its analytical power, ULaMDyn is designed for accessibility and scalability. Researchers can run the tool on standard laptops or integrate it into high-performance computing workflows for larger systems. Its modular architecture supports customization, making it adaptable to a wide range of molecular systems and research objectives.
Looking ahead, ULaMDyn will continue to evolve with new features, including additional molecular descriptors and advanced clustering methods, broadening its applicability to even more complex systems. By bridging machine learning and molecular dynamics, ULaMDyn represents a significant step forward in our ability to understand and harness excited-state phenomena.
For detailed insights into ULaMDyn’s capabilities and applications, explore the full article in Digital Discovery or visit the ULaMDyn website and its official GitLab repository. Whether you are tackling photochemical reactions or materials design, ULaMDyn is poised to be an indispensable resource in your computational toolkit.
MB
Reference
[1] M. Pinheiro, Jr., M. O. Bispo, R. S. Mattos, M. Telles do Casal, B. C. Garain, J. M. Toldo, S. Mukherjee, M. Barbatti, ULaMDyn: Enhancing Excited-State Dynamics Analysis Through Streamlined Unsupervised Learning, Digit. Discov. (2025). DOI: 10.1039/D4DD00374H